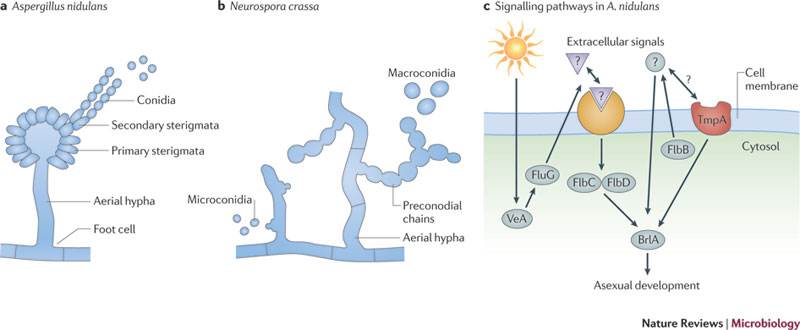
Transcription factors are proteins that bind to DNA to promote or suppress protein production. Since almost all diseases involve disruption of the protein-production process, transcription factors are promising biological targets for drugs — and could even serve as drugs themselves.
But there are likely thousands of transcription factors in humans, each of which might bind to the genome at tens of thousands of different locations. Previously, there was no cost-effective way to figure out exactly where transcription factors bind — which exact DNA letters in a given stretch of genome each of them attaches to. Biologists thus relied on approximate methods to identify the general vicinity of binding sites.
In the August issue of the online journal PLoS Computational Biology, a team of researchers from MIT’s Computer Science and Artificial Intelligence Laboratory presented a new analytic technique that identifies binding sites with much greater accuracy. As a consequence, the researchers were able to infer previously unknown relationships among transcription factors, which could provide clues to the roles they play in biological processes.
The researchers initially tested their technique on two sets of experimental data, which they say represent both “relatively easy and difficult cases” for analysis. In the easy case, their new technique identified the precise locations at which transcription factors bound to the genome with more than 90 percent accuracy, while the accuracy of existing techniques was about 10 percent or less. In the difficult case, the new method was more than 55 percent accurate, compared to about 5 percent for existing techniques.
The leading method for determining how transcription factors behave in living cells is to chop up the DNA from millions of cells and use protein antibodies to extract the fragments that have a particular transcription factor attached to them. While the DNA sequence that a transcription factor binds to consists of only about six to 12 DNA letters, the fragment extracted by the antibody could be a couple of hundred letters long. Sequencing the fragments can determine where in the genome they came from, but it offers little information about where on the fragment the transcription factor is attached.
Feedback loop
David Gifford, a professor of electrical engineering and computer science and director of the Computational Genomics Group, his graduate student Yuchun Guo, and Shaun Mahony, a research scientist in the group, developed a new algorithm for analyzing millions of experimentally identified fragments and inferring the precise locations at which transcription factors bind to them.
Previous methods would compare the fragments to try to identify sequences they had in common. But that’s just the first step in the MIT researchers’ method. They then use that initial, rough guess about common sequences to predict where, throughout the entire genome, the transcription factor would bind, then compare those predictions to the experimental data on where the factor actually did bind. On the basis of that comparison, they then refine their estimate of the specific binding sequence and repeat the whole process.
“We iterate between estimating where proteins bind and using that information to discover the sequences that they bind to,” Gifford says, “and then we go backward and use the sequences they bind to to improve the estimate of where they’re binding.”
But determining transcription factors’ precise binding sites is just the first step in understanding their role in protein production. For a single transcription factor, that role can vary according to both the type of cell in which it’s active and its interactions with other transcription factors. It’s the second of these elements that the MIT researchers are shedding light on, by identifying spatial relationships between binding locations that imply a functional relationship between the corresponding transcription factors.
The genome’s language
That approach, Gifford says, is similar to the statistical analysis of language, which artificial-intelligence researchers have used to build language-interpreting computer systems. Indeed, Gifford says, the sequences that transcription factors bind to can be thought of as words and their spacing as the “syntax” of the genome.
“If you did an analysis of the English language, you would find a lot of relationships between words that were highly significant, because they co-occur,” Gifford says. “You would not necessarily understand from the analysis what their meaning was, but you would know that they were highly significant and did carry meaning.” The same is true of the DNA “words” that constitute the transcription-factor binding sites. “If you look at a null model, which would posit random occurrence of words, then you ask how unlikely it is that you would see these things together,” Gifford says. “And we’re testing everything against a random model.”
The MIT researchers’ analysis identified a handful of relationships between transcription factors that were already known, but it also identified 390 more statistically significant relationships between binding sites. Some of those may be red herrings, but many of them could turn out to indicate previously unsuspected relationships between transcription factors, which could help biologists unravel the mysteries of genetic expression.
“I think it’s beautiful work. I really like it,” says Michael Snyder, a professor of genetics at Stanford University and one of the lead investigators on the National Human Genome Research Institute’s massive ENCODE project to characterize all the functional elements of the human genome. Just last week, Nature published a paper called “Architecture of the human regulatory network derived from ENCODE data,” which had dozens of authors, including Snyder and a number of other ENCODE researchers.
“In much higher detail than we had done in our paper, Gifford’s work could really let us understand much better about how these proteins are functioning together,” Snyder says. “This is really going to be critical for us to understand the basic biological pathways for how you develop a human being and, of course, ultimately, for what goes wrong in human disease.”
 ASUS X101CH-EU17-WT 10.1-Inch Netbook (White)
ASUS X101CH-EU17-WT 10.1-Inch Netbook (White)ASUS X101CH-EU17-WT EeePC Netbook / 10.1″ WSVGA (1024 x 600) / Intel Atom N2600 (1.6GHz) / 1GB DDR3 / Intel UMA / 350GB 5400RPM / …
 Apple iBook G4, 1.33 GHz, 768MB RAM, 40 GB Hard Drive, Internal Combo Drive, 56k Modem
Apple iBook G4, 1.33 GHz, 768MB RAM, 40 GB Hard Drive, Internal Combo Drive, 56k ModemRAM of 768MB ; 40 GB internal drive internal Combo Drive ; internal 56k modem Airport Extreme and Bluetooth installed ; 12.1″ TFT …